Department of Biotechnology, School of Engineering & Technology, Sharda University, Greater Noida, Uttar Pradesh-201310, India. E-mail address: saurabh.jha@sharda.ac.in
Ojective: In-silico methods to find and characterize the ligands against the active site of tau protein which could assist in the therapeutics of Alzheimer’s disease.
Methods: The aid of various bioinformatic tools such as phylogenetic analysis, homology modeling, and active site prediction led to the molecular docking analysis of the major malefactor for Alzheimer’s disease ‘microtubuleassociated tau protein’. A three-dimensional structure of microtubule-related tau protein was created, and the Ramachandran plot was acquired for quality appraisal.
Results: Procheck showed 62.95 of residues in the most preferred region with 20% residues in the additional allowed region and 5.7 % in the disallowed region of microtubule-associated tau protein. Screenings of the particles were done dependent on Lipinski’s standard of five.
Conclusion: Genistein, Hesperidin, and epigallocatechin-3 are the potential ligands in regulating microtubule-related tau protein and Epigallocatechin-3 gallate is the most potent among them and the most elevated negative free vitality of official with the maximum interacting surface territory throughout docking studies.
Keywords: Alzheimer’s disease, Docking Ligand, Tau Protein, Dementia
Alzheimer’s disease is a dreadful brain disorder that is influenced by environmental and genetic factors. The disease affects many people worldwide and various studies are undergoing to determine the exact cause and treatment of the disease. In a recently published study, 11 markers for AD were identified in 10 loci using genome-wide complex trait analysis to examine >2 million SNP’s for 10,922 individuals.[1] AD is the most common type of dementia since the worldwide rate of AD is 24 to 34 million people.[2] Age is the most vital factor which influences AD progression.[3],[4] Additionally, factors like hypertension, estrogen supplements[5], smoking[6],[7], stroke, heart disease, depression, arthritis, and diabetes[7] and some minor lifestyle choices also might involve the risk of AD. The positive choices which could decrease the risk of AD include intellectual stimulation[8], exercise[9], and a Mediterranean diet centered with vegetables and fruits including fish and red meat occasionally.[5],[10] AD therapeutics begins with the determination of its causative factors which is done with an electron microscopic view of abnormal filaments formed in the brain. The analysis of these abnormal deposits assists in AD therapeutics and disease classification. It is yet unclear what are the functions and clear cause of these deposits but two major components that affect the treatment of AD are amyloid-β as the major plaque component and tau as the major tangle component. Tau deposits are also seen in other neurodegenerative disorders which include Pick’sdisease.[11] The main component of the tangles is tau protein, and it can bind and stabilize the cell’s internal skeleton called microtubules. Tau proteins are a member of family microtubule-associated proteins that are majorly expressed in neurons where it has a crucial role in the assembly of tubulin monomers into microtubules to make up for the neuronal microtubule network.[12],[13] Tau protein is a phosphoprotein that has 79 potential serine (Ser) and threonine (Thr) phosphorylation sites on the longest observed tau isoform. The phosphorylation has been reported on nearly about 30 of these sites in normal tau proteins. Through various studies done over a while, the tau protein is found to be the major constituent of intraneuronal and glial fibrillar lesions in Alzheimer’s disease and thus numerous neurodegenerative disorders are referred to as ‘tauopathies’. A direct correlation has been found between the progressive involvement of the neocortical areas and the increasing sensitivity and severity of dementia suggesting that the tau proteins are a reliable marker of neurodegenerative processes. [14] Many pharmacological treatments have been proposed and used for neurodegenerative disorders, but the treatments are symptomatic and are not able to bring a change in the progression of neurodegenerative disorders. The current treatment methods are less satisfactory and might lead to serious side effects thereby the goal of this research is to develop a potential inhibitor of tau protein that also has the potential to be used in the therapy of AD.
Software required
The amino acid sequence of tau protein was downloaded from NCBI Database using BLAST, and protein function elucidation was done using the Interproscan server. The phylogenetic relationship and multiple sequences were analyzed using Muscle software and the ProtParam tool was used to find the physicochemical properties. The homology modeling was done to study the 3D structure of microtubuleassociated tau protein. PDB ID 2MZ7 was used to recover proteins for tau protein and the protein structure was downloaded using the Swiss model further modeled structure was visualized using the PyMol. Auxiliary assessment and stereochemical examination were performed using RAMPAGE. The ligand molecules were retrieved from the PubChem compound database and the visualization of the 3D image of the compound was done using PyMOL. The likeliness of the drug was retrieved using Lipinski’s filter. The active site of tau protein was predicted using CastP, further the preparation of protein and ligand molecule was done using the Chimera tool. Finally, the docking of ligand and the protein was done using Auto Dock 4.2.6 and predict the possible protein-ligand interactions.
Docking Methodology
Retrieval of tau protein and their function recognition
The amino acid sequence of tau protein was retrieved from the NCBI database and used for homology search using the basic local alignment search tool (BLAST). Proteins functions elucidation was done using Interproscan server (http://www.ebi.ac.uk/interpro/search/sequence/)
Phylogenetic relationship and physicochemical properties
Muscle software (https://www.ebi.ac.uk/Tools/msa/muscle/) was used for multiple sequence analysis and a phylogenetic tree was constructed using muscle software based on the neighbor-joining plot without distance correction. The ProtParam tool (https://web.expasy.org/protparam/) was used to predict physicochemical properties. The parameters computed by ProtParam included the molecular weight, PI, aliphatic index, and GRAVY.
Homology modeling and visualization of the 3D structure of Microtubule-associated Tau protein
Homology modeling was utilized to decide the 3D structure of microtubule-associated Tau protein. Formats with PDB ID 2MZ7 were recovered for Tau proteins from PDB. The protein structure forecast server Swissmodel (https://swissmodel.expasy.org/) was utilized for homology. Further, the modeled structure was visualized using PyMOL. Auxiliary assessment and stereochemical examination were performed utilizing RAMPAGE. (http://www.mordred.bioc.cam.ac.uk/~rapper/rampage.php).
Ligand optimization
Ligand molecules Genistein, Hesperidin, and Epigallocatechin- 3 gallate along with their physical and chemical properties were retrieved from the PubChem compound database. 3-D image of ligand was downloaded in SDF file and was a visualization of the molecular structure of the compound was done using PyMOL. (https://pubchem.ncbi.nlm.nih.gov/)
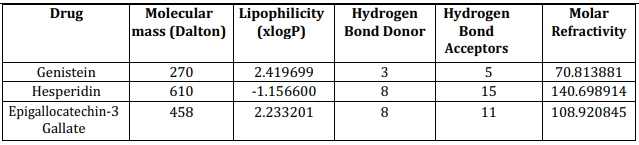
Lipinski’s filter analysis of screen drug
To retrieve the information about the drug-likeness of drug, online tool Lipinski’s filter (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) was used. Lipinski’s rule of five (or Lipinski’s rule) help us to identify the possibility of success or failure due to drug-likeness for molecules fulfilling with two or more of the following rules: (a) Molecular mass should be less than 500 Dalton, high lipophilicity (expressed as LogP less than 5), less than 5 hydrogen bond donors, less than 10 hydrogen bond acceptors, molar refractivity should be between 40-130.
Active site prediction
CastP (http://sts.bioe.uic.edu/castp/index.html?1ycs) was used to predict the active sites of the protein. PDB file of protein was uploaded to the server and it showed the ligand-binding sites present in protein and the site with maximum surface area and maximum surface volume was selected and all the amino acid residues involved in binding with ligands were retrieved.
Preparation of protein and ligand molecule
Preparation of protein includes the expansion of polar hydrogen atoms, expansion of charge, and expulsion of any various structures from the protein atom by Chimera tool, though ligand preparation includes the addition of charge.
Molecular docking analysis
Prepared and optimized structures of ligands and proteins were ultimately used for molecular docking using Auto Dock 4.2.6 for predicting the possible protein-ligand interactions and the results that include the understanding of the association that involves H-bonding and hydrophobic interactions were analyzed using LIGPLOT1.4.5, a program to generate schematic diagrams of protein-ligand interactions.
Retrieval of amyloid-beta precursor protein (AβPP)
A BLAST search was done based on the functional domain sequence of a well-characterized gene or protein of interest. There are 6 isoforms of tau.[11] These isoforms differ in 3 or 4 tubulin-binding domains within the C-terminal region.[12] The Interproscan result was able to successfully hunt 2 isoforms based on families and domains that were obtained in the result. The Interproscan results also demonstrated that the homologous protein for tau were the members of the Microtubule-associated protein (IPR027324), MAP2/MAP24/TAU, and (IPR001084), MAP_tubulin-bd_rpt accordingly. The results are shown in Figure 1.
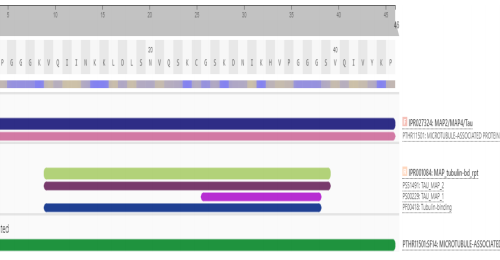
Figure 1: Interproscan result for tau protein
Phylogenetic relationship and physicochemical properties
Muscle software was used for running the multiple sequence analysis and it was discovered that the amino acid residues were conserved in the isoform sequences as well. The results of the multiple alignment sequence are shown in the Figure-2a and 2b. Table 1 & 2 demonstrates the physiochemical properties of the concerned protein using the ProtParam tool. The molecular weight of the tau protein was found to be 4882.66 Da and an isoelectric point of tau protein was 40.63 which pointed towards the charge present on the protein and also the GRAVY index was found to be -0.698.
Figure 2: (a) Multiple sequence alignment of all tau protein isoforms and (b) Tree generation for tau protein using neighbor-joining plot without distance correction
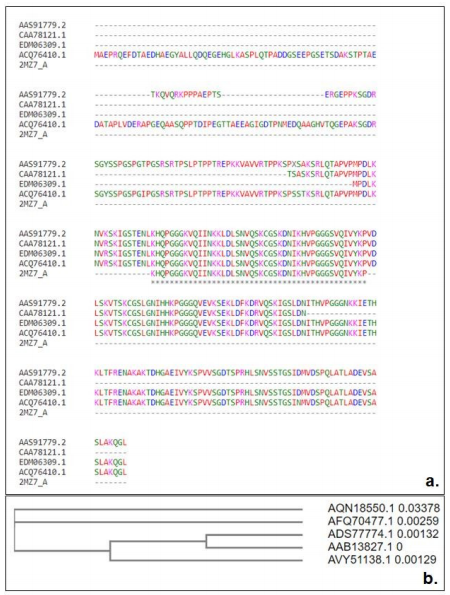
Table 1: Physiochemical properties of microtubule-associated tau protein

Table 2: Physiochemical properties of ligands

Homology modeling, visualization, and quality assessment of the 3D structure of AβPP
The validation of the final model was done by using the Ramachandran plot computed with PROCHECK. This program helps in checking the detailed structure residue-by-residue stereo-chemical quality of a protein structure and also assess in the stereochemical assessment of the model and it is accessible in Table 3
The criteria for analysis of stereochemistry of the model include:
Main chain conformation in the acceptable region of the Ramachandran plot, Planar peptide bonds, Sidechain conformations that correspond to those in the rotamer library, Hydrogen bonding of polar atoms if they are buried, No bad atom-atom contacts, No holes inside the structure.[15]
Ramachandran Plot:
A Ramachandran plot also called a Ramachandran map or a Ramachandran diagram developed by Gopalasamudram Narayana Ramachandran and Viswanathan Sasisekharan is a way to visualize dihedral angles Ψ and Φ of amino acid residues in protein structure. The plot exhibits the possible conformations of Φ and Ψ angles for a polypeptide thereby, the Ramachandran plot is a useful way of assessing the stereochemical quality of the targeted protein structure and the main Ramachandran plot was developed using PROCHECK and the results indicated that tau protein had 247 (94.3%) residues in the favored region against (~98.0% expected), 10 (3.8%) residues in the allowed region against (~2.0% expected) and 5 (1.9% residues in outlier region as shown in Figure 3.
Table 3: Homology search defining biomolecules, score, identity, and E value.
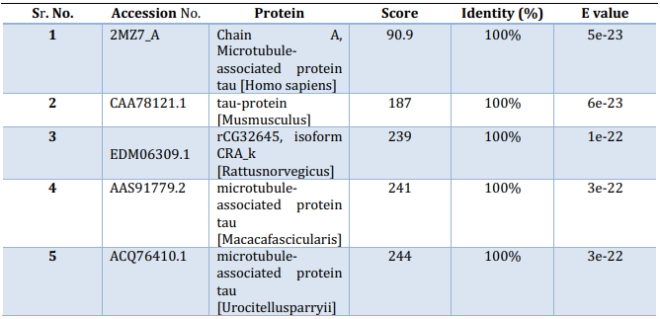
Figure 3:
Three-dimensional (3D) structure, Ramachandran plot, and active site of tau protein. (a) Ramachandran plot obtained for quality assessment, (b) 3D structure of tau protein and active sites in tau protein (c) Ligand binding site in tau protein

Ligand Optimization
The 3-D structure of the three ligands Genistein, Hesperidin, and Epigallocatechin-3 gallate were visualized by using PyMOL and the displayed in Figure 4 given below.
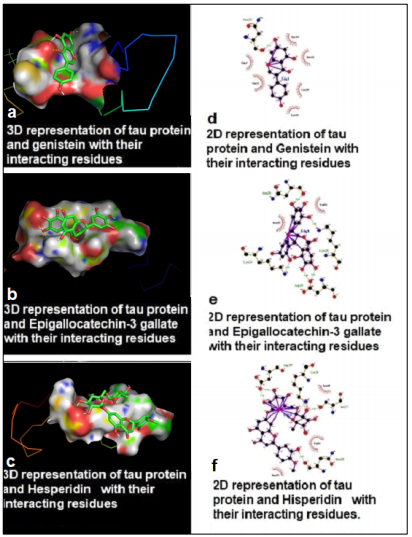
Figure 4:
Binding site of tau protein with selected compounds along with its known stimulatory active site. (a) Three-dimensional (3D) representation of tau protein and Genistein with their interacting residues, (b) 3D representation of tau protein and Epigallocatechin-3 gallate with their interacting residues, (c) 3D representation of tau protein and Hesperidin with their interacting residues, (d) Two-dimensional (2D) representation of tau protein and Genistein with their interacting residues, (e) 2D representation of tau protein and Epigallocatechin-3 gallate with their interacting residues, (f) 2D representation of tau protein and Hesperidin with their interacting residues
Lipinski’s filter analysis of screened drugs
In modern drug discovery, various screening methods with high throughputs have become a more indispensable approach to identify the drug leads. Lipinski’s rule of five is the most popular and sought-out method to find the drug-likeness. It was reported that 90% of oral bioavailability of non-CNS drugs had a PSA below 120 Å2, and the criterion was dropped to 80 Å2 for CNS drugs.[15] The Lipinski’s rule of five is as followed: Molecule with a molecular mass less than 500 Da, No more than 5 hydrogen bond donors, No more than 10 hydrogen bond acceptors, Octanol-water partition coefficient log P not greater than 5, Molar refractivity should be between 40-130. The results of the Lipinski rule of five are shown in Figure 5 and Table 4 below.
Figure 5: Drug likeness prediction using Lipinski’s filter analysis

Table 4: Drug likeness prediction using Lipinski’s filter analysis
Active site prediction
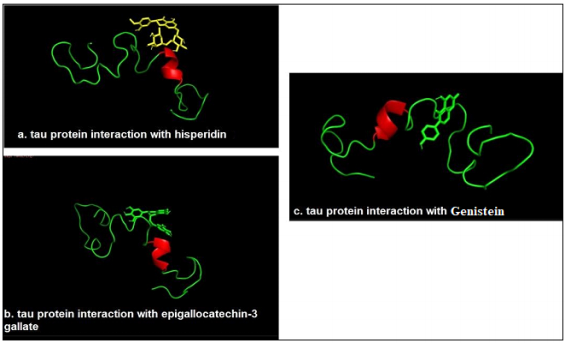
The identification of active sites of proteins and DNA are linked with the structural pockets and cavities that were found out by using the CastP server. This server is capable of predicting the possible active sites from the 3D atomic coordinates of the protein of interest. Tau protein in this particular study involved the ligand-binding site and site volume. The PDB file was uploaded and the ligand-binding sites were seen for the targeted protein. The site of maximum surface area was used and is shown in Figure 6, and the amino acids residues participating in the binding with the ligands separately for all the ligand molecules were retrieved.
Figure 6: Docking study of tau protein with selected biomolecules (a) tau protein interaction with Hesperidin (b) tau protein interaction with Epigallocatechin-3 gallate (c) tau protein interaction with Genistein
Preparation of protein and ligand molecules
Chimera tool was used to analyze the target protein i.e. expansion of polar hydrogen atoms, expansion of charge, and removal of any miscellaneous structures from the protein molecule by AutoDock 4.2.6, and the ligand preparation involved addition of charge done by Chimera tool.
Molecular Docking Analysis
The binding of the ligand molecules with the protein molecule (receptor) was analyzed using AutoDock 4.2.6 to find the possible protein-ligand interaction by calculating its hydrogen bond and ionic interactions and configuration of the whole ligand molecule to obtain the minimum energy structure. Docking was performed for three different ligands separately and the parameters used for the molecular docking are accessible at http://autodock.scripps.edu/resources/parameters. The interaction between the modeled stable tau protein structure and ligand molecules is shown in Table 5. Among all of them, the Epigallocatechin -3 gallate has the highest energy and affinity towards the target protein. The results of the docking are shown in Table 6.
Table 5: Tau protein known binding site and selected compounds interacting residues
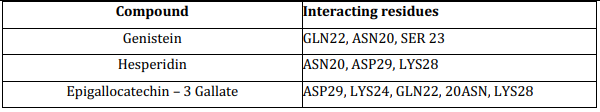
Table 6: Docking result showing the binding energy and number of rotatable bonds
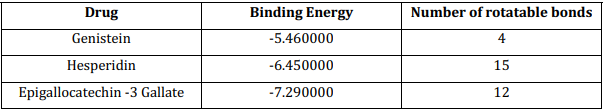
The abnormal functioning of tau protein which causes the formation of tangles should be controlled by blocking the active sites of tau protein i.e. target protein and the active sites are found using the CastP and the 3D structure of the active site was viewed using the tools like Chimera tool and these ligand molecules used in the study are used for targeting these active sites of the targeted protein. The studies in the analysis of tau protein tend to find a therapeutic cure for AD and the same purpose the three bimolecular drugs Genistein, Hesperidin, and Epigallocatechin- 3 gallate were used to find the best suitable candidate with the best ligation with the tau protein and the results show a successful binding of two drugs out of which Epigallocatechin- 3 gallate with a molecular mass of 458 Da and 8 Hydrogen bond donor and 11 hydrogen bond acceptors and a molar refractivity of 108.920845 shows binding energy of – 7.290000. The most vital interactions which involve a ligand and receptor’s active site are hydrogen bonding and ionic interactions. These suggest that the potential ligand should have stronger hydrogen and ionic interaction with the moieties of the binding sites.[16],[17]
The study focused on finding the potential of biomolecules Genistein, Hesperidin, and Epigallocatechin -3 gallate for the therapy of AD. The sequence and structure analysis of tau protein was done using in-silico methods and bioinformatics software. The result of the molecular docking study conferred that the drug Epigallocatechin- 3 gallate was the most effective in binding with the tau protein based on the highest negative free energy of binding, minimum inhibition constant Ki, and the maximum interacting surface area among the three given biomolecules. The capability of Epigallocatechin- 3 gallate is not limited to this study but was effectively used to validate in vitro and vivo prosurvival outcomes in AD models as well as in clinical scenarios. The study aims at providing a glance at the broad screening of these inhibitors of tau protein and they can be further implemented in the designing of therapeutics for AD.
The authors declare that there are no conflicts of interest relevant to this article.
References
- 1.Ridge PG, Mukherjee S, Crane PK, Kauwe JS; Alzheimer’s Disease Genetics Consortium. Alzheimer’s disease: analyzing the missing heritability. PLoS One. 2013 Nov 7;8(11):e79771. [Google Scholar] [PubMed]
- 2.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007 Jul;3(3):186-91. [Google Scholar] [PubMed]
- 3.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011 Mar 19;377(9770):1019-31. [Google Scholar] [PubMed]
- 4.Herrup K. Reimagining Alzheimer’s disease-an age-based hypothesis. J Neurosci. 2010 Dec 15;30(50):16755- 62. [Google Scholar] [PubMed]
- 5.Patterson C, Feightner JW, Garcia A, Hsiung GY, MacKnight C, Sadovnick AD. Diagnosis and treatment of dementia: 1. Risk assessment and primary prevention of Alzheimer disease. CMAJ. 2008 Feb 26;178(5):548- 56. [Google Scholar] [PubMed]
- 6.Cataldo JK, Prochaska JJ, Glantz SA. Cigarette smoking is a risk factor for Alzheimer’s Disease: an analysis controlling for tobacco industry affiliation. J Alzheimers Dis. 2010;19(2):465-80. [Google Scholar] [PubMed]
- 7.Almeida OP, Hulse GK, Lawrence D, Flicker L. Smoking as a risk factor for Alzheimer’s disease: contrasting evidence from a systematic review of case-control and cohort studies. Addiction. 2002 Jan;97(1):15-28. [Google Scholar] [PubMed]
- 8.Wang HX, Karp A, Winblad B, Fratiglioni L. Late-life engagement in social and leisure activities is associated with a decreased risk of dementia: a longitudinal study from the Kungsholmen project. Am J Epidemiol. 2002 Jun 15;155(12):1081-7. [Google Scholar] [PubMed]
- 9.Podewils LJ, Guallar E, Kuller LH, Fried LP, Lopez OL, Carlson M, Lyketsos CG. Physical activity, APOE genotype, and dementia risk: findings from the Cardiovascular Health Cognition Study. Am J Epidemiol. 2005 Apr 1;161(7):639-51. [Google Scholar] [PubMed]
- 10.Scarmeas N, Stern Y, Mayeux R, Luchsinger JA. Mediterranean diet, Alzheimer disease, and vascular mediation. Arch Neurol. 2006 Dec;63(12):1709-17. [Google Scholar] [PubMed]
- 11.Lei P, Ayton S, Finkelstein DI, Adlard PA, Masters CL, Bush AI. Tau protein: relevance to Parkinson’s disease. Int J Biochem Cell Biol. 2010 Nov;42(11):1775-8. [Google Scholar] [PubMed]
- 12.Kolarova M, García-Sierra F, Bartos A, Ricny J, Ripova D. Structure and pathology of tau protein in Alzheimer disease. Int J Alzheimers Dis. 2012;2012:731526. [Google Scholar] [PubMed]
- 13.Billingsley ML, Kincaid RL. Regulated phosphorylation and dephosphorylation of tau protein: effects on microtubule interaction, intracellular trafficking and neurodegeneration. Biochem J. 1997 May 1;323 (Pt 3):577-91. [Google Scholar] [PubMed]
- 14.Solfrizzi V, Scafato E, Seripa D, Lozupone M, Imbimbo BP, D’Amato A, et al. Reversible Cognitive Frailty, Dementia, and All-Cause Mortality. The Italian Longitudinal Study on Aging. J Am Med Dir Assoc. 2017 Jan;18(1):89.e1-89.e8. doi: 10.1016/j.jamda.2016.10.012. PMID: 28012505. [Google Scholar] [PubMed]
- 15.Kumari R, Chaudhary A, Mani A. Casuarictin: A new herbal drug molecule for Alzheimer’s disease as inhibitor of presenilin stabilization factor like protein. Heliyon. 2020 Nov 21;6(11):e05546. [Google Scholar] [PubMed]
- 16.Kelder J, Grootenhuis PD, Bayada DM, Delbressine LP, Ploemen JP. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res. 1999 Oct;16(10):1514-9. [Google Scholar] [PubMed]
- 17.Patterson C, Feightner J, Garcia A, MacKnight C. General risk factors for dementia: a systematic evidence review. Alzheimers Dement. 2007 Oct;3(4):341-7. [Google Scholar] [PubMed]

